April 2016
Seals from a new perspective
Raven Benko, undergraduate student
29 April 2016
This last month I have been fortunate enough to study seals in a new environment – an experience that has been exciting, demanding, and breathtaking all at once. As part of a feasibility study being run by Dr. Acevedo-Gutierrez and his newest graduate student, Madelyn Voelker, a few lucky undergrads like myself have been helping out in a quest for seal scat! It may not sound very glamorous, but sampling and analyzing animal feces is a commonly used noninvasive method for monitoring marine species. There are even trained dogs across the world that are used to smell out fecal matter from marine and other animals so that such samples can be collected. Luckily for us, due to harbor seals’ haul-out behaviors, we only need to search frequented haul-out sites for scat samples, and we’ve been having some luck!
Check out this video about Tucker the poop-sniffing dog in the University of Washington’s Conservational Biology Department.
Scat can tell you a lot of information about an animal, including its sex, diet, and overall health. For our study, we are performing a dietary analysis in order to better understand what the population is eating, and possibly whether or not there is a difference in diet between individuals. What does this information have to do with the overall state of the Salish Sea? The answer is salmon. In our region, as most already know, the fate of our native salmon populations is of utmost concern to local citizens, First Nations and Tribal members, resident orcas, and more. Many people want to know if our abundant harbor seal populations are largely relying on salmon for food just like our charismatic killer whales and us humans. Our study, along with other similar studies in the Pacific Northwest Area, will hopefully shed some light on this subject and see just how much an influence harbor seals have on salmon populations.
My first time on our research vessel we traveled to Seal Rocks and the Penn Cove Shellfish Farm but had no luck finding any seal scat. It turns out that seal scat is pretty hard to find. If the weather is too cold for seals to haul-out or there has been a lot of wave activity that washes the poop off the haul-out site, you will be hard-pressed to find any fresh samples. However, as we progress into spring, the study has been finding more and more poop. Just last Monday I helped Madelyn collect 10 samples off the floating shellfish docks at Penn Cove. The floating docks were covered with seals and these individuals seemed to be very accustomed to human interaction. A few of them even let us step on to the dock before fleeing the structure. Looking back at my post from last week it appears that there is great variation in the amount of pedestrian disturbance harbor seals will tolerate. Some habituation appears to have occurred with the population of seals in Penn Cove, as they allow humans to approach up to 10 meters or less before fleeing a haul-out site.

April 25, 2016: Me, in a full Mustang suit.

April 25, 2016: View from Seal Rocks.
Photo: R. Benko
Along with poop and harbor seals, I have seen many other interesting things on these trips. On Monday we saw a California sea lion sunning itself on a large buoy. The trip before that I saw what appeared to be an aborted harbor seal fetus on Seal Rocks. The pup definitely looked premature at the very least. Overall it has been an exciting project to participate in and I am looking forward to future trips!

April 15, 2016: Dead harbor seal on Seal Rocks.
Photo: R. Benko
Use of R in the Whatcom Creek Project Lab
Daniel%20Woodrich, undergraduate student
29 April 2016
Ecology isn’t all about fieldwork; making the schedule to get out there, analyzing data, running stats and producing visualizations all have to be done in the lab, and this year they have been primarily accomplished by using R studio.

R has gained popularity for data manipulation in the field of biology in the last few years for a few reasons.
1-Traceability. The convention when using R is to work in a source window (see image below). Scripts written in the source window can be “bumped down” selectively to the console window to be run, and this allows the user to be able to try different possibilities, and follow a whole process from the start to the finish without actually running it in the console (where code actually does its function).
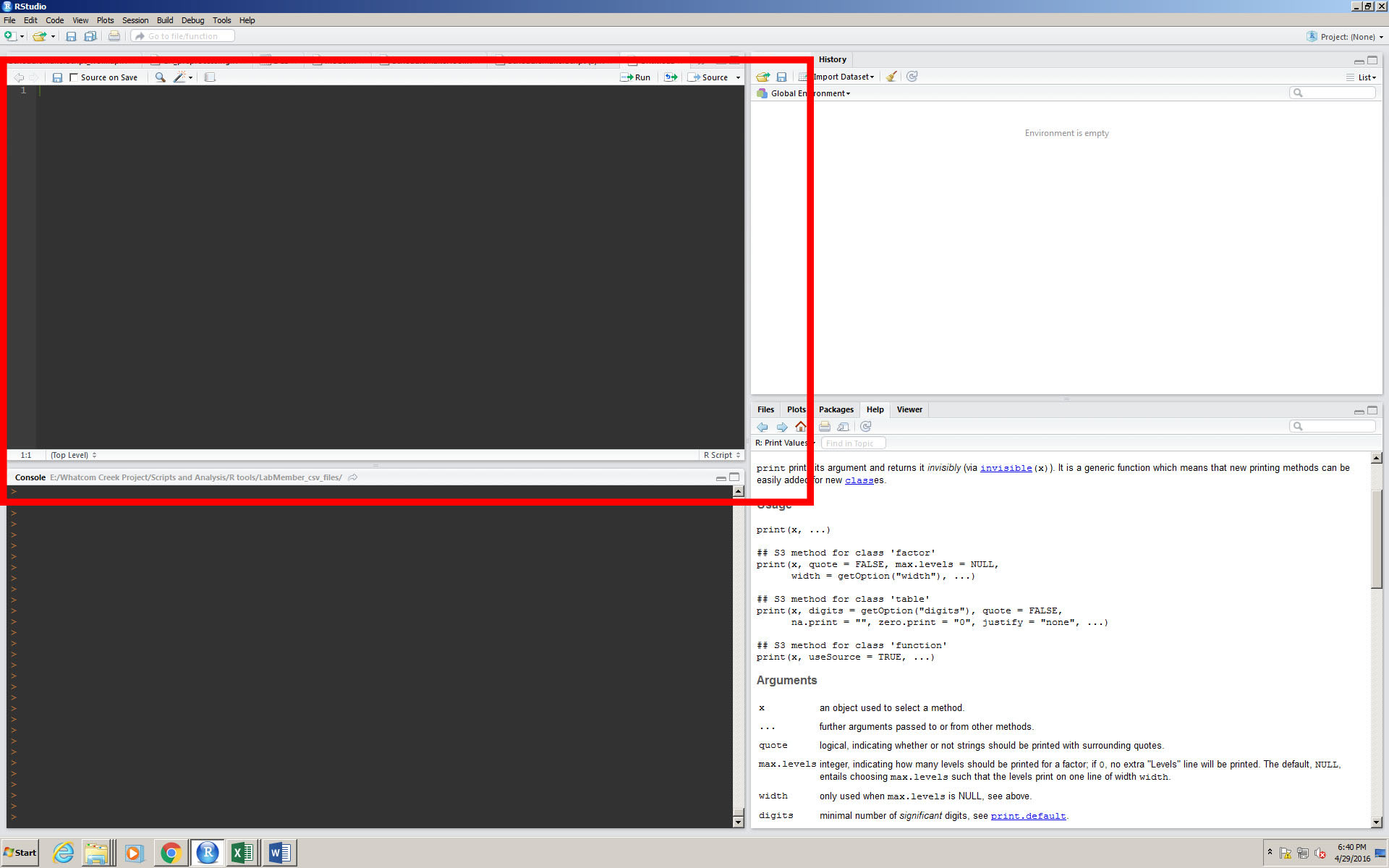
2-Transferability. R studio is used by scientists everywhere, and complex functions that serve specific purposes can be useful to others in your field. Sharing code is a tenant of R programmers everywhere, and is passed around from person to person, or on sites like GitHub.
This year, I used R studio to create a new solution to one of the consistent time consuming jobs that myself and previous managers have encountered: making schedules. Since the project’s inception, it has been procedure to go out to the creek during slack tide to observer, based on a recommendation by the hatchery manager that more fish aggregate during the slack periods. Because of this, schedules vary in their timing, and frequency: since we only observe in daylight conditions, we have fewer observations available to us in the winter. An example schedule can be seen below:
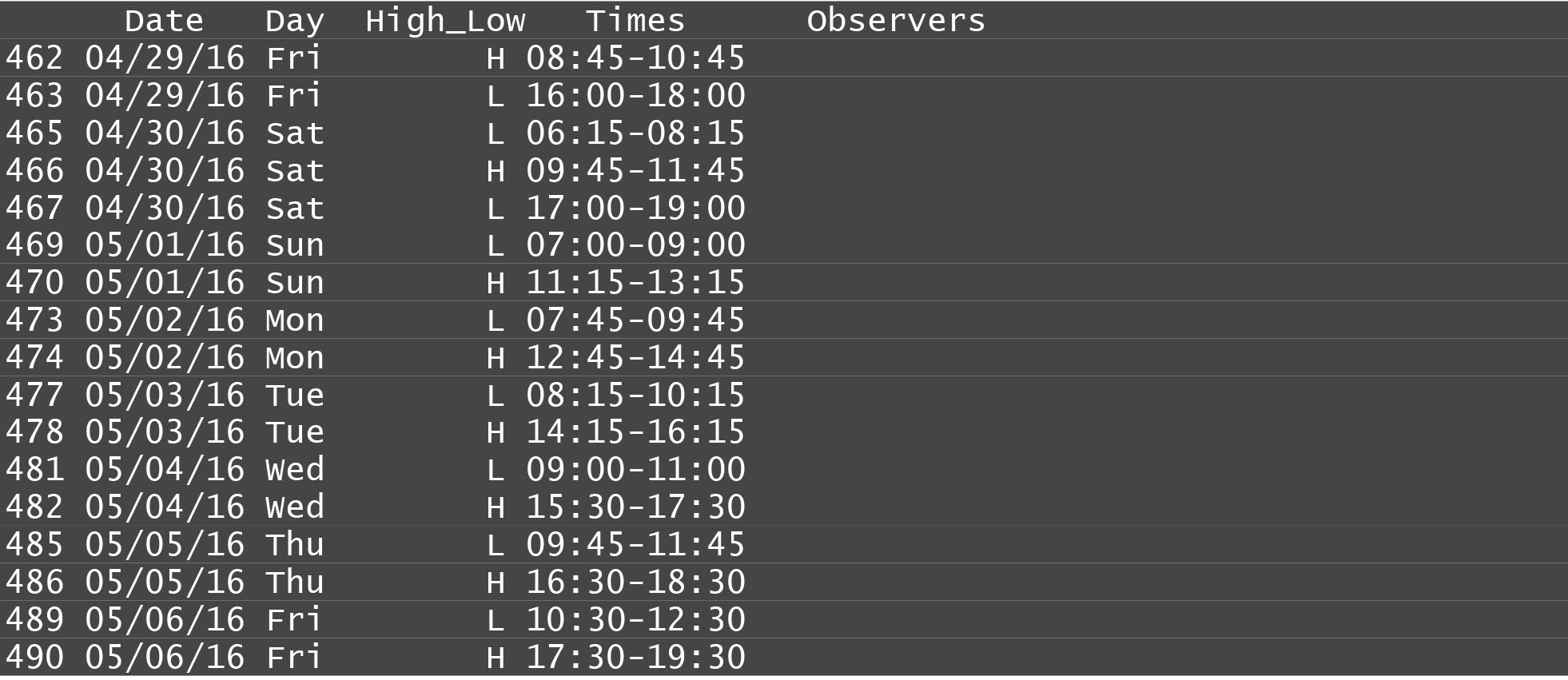
These schedules used to be made from hand in excel, and could take up to an hour every two weeks to assemble. The fact that data on both tide and sunrise and sunset times were available publically for download online at NOAA enabled this tool to exist.
I decided to create a program, rather than a source script, to accomplish this function. Programs give users a series of prompts, and don’t require them to interpret and modify code. This is advantageous both for simplicities sake, and the fact that it enables any user to use tools you develop.
To operate the program, the user simply navigates to “Run all” (referring to the source window) and types responses in the console:
Here, the user wants their schedule to start from today, 04/29/16.
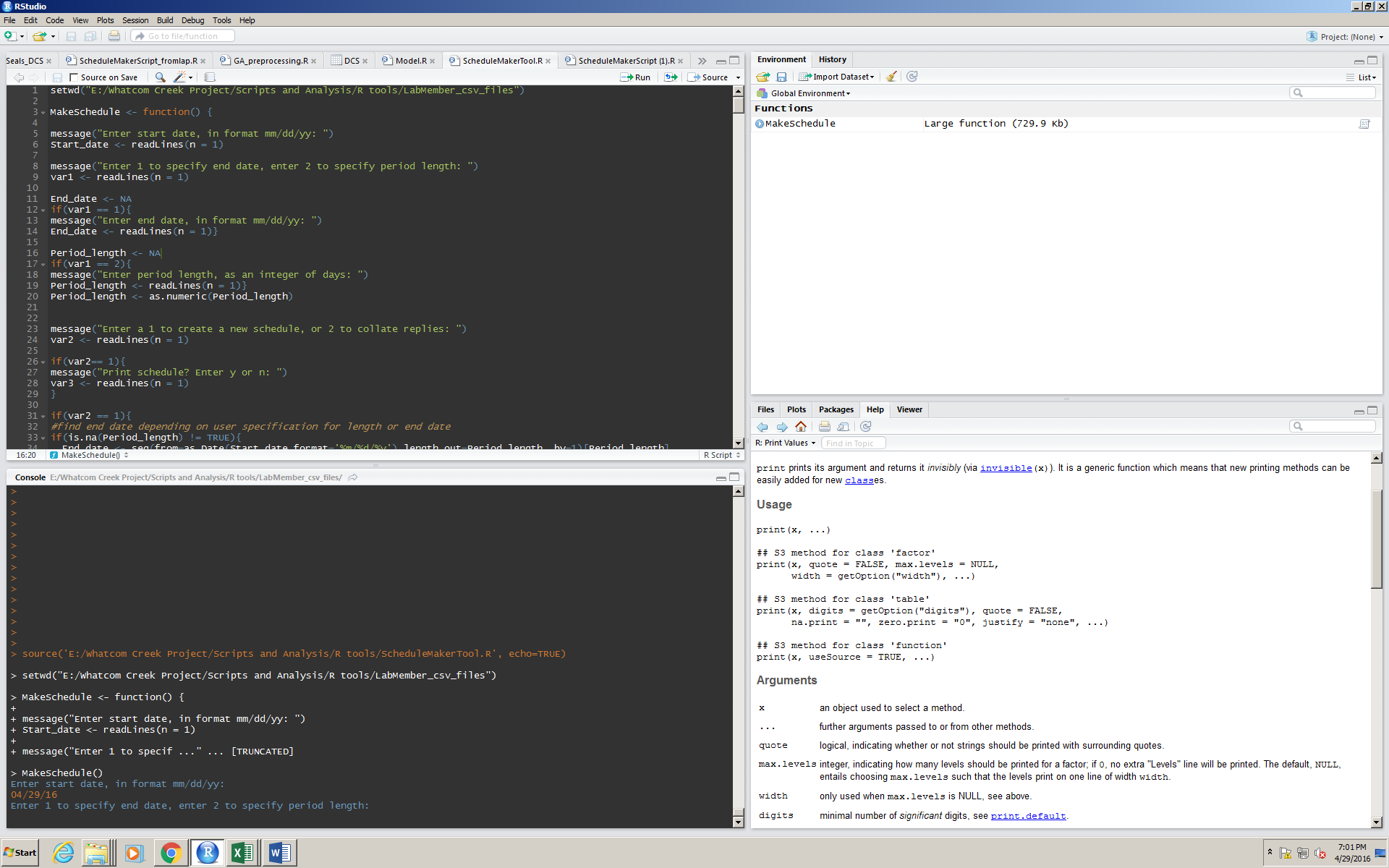
The user can pick whether they want a schedule that goes to a certain date, or go for a certain number of days by inputting a 1 or a 2.

After picking an end date, the program asks if the user wants to print a new schedule, or paste together the email responses from the lab members for availability (step 2 of the scheduling process). If you want to save the schedule in order to send it out to the lab members (step 1 of the scheduling process), the user can enter a y, and the program will print where the new file is saved to the console.
The program will behave the same way if you choose to enter a period length instead of an end day.

R, or programs similar to it (such as Python and MATLAB) are used in just about every research lab in the country. Aside from this program, we have used R to check for errors in our dataset, create visualizations, summarize and analyze our data, and are in the process of generating a Generalized Linear Mixed Model to predict the occurrence of seals in our study site using environmental variables. Programming can be used in versatile ways as a time saver, as seen here, or as an essential tool to manipulate and understand your data.
Track what I have been doing with programming and data analysis relevant to our lab on my GitHub page.
Code is most useful if it can be communicated, shared, and accessible for others!