April 2017
Microsatellites analysis
Madelyn Voelker, graduate student
1 April 2017
Microsatellites are non-coding regions of DNA that have a large number of repeats of a short pattern. Due to the highly repetitive nature of microsatellites, mistakes are relatively common in those sections when replicating, resulting in the addition or deletion of a repeat. Further, because microsatellites are non-coding regions, there is no selective pressure to weed out any of them. This fact often results in a large number of alleles, each with a different number of repeats and therefore different lengths, in microsatellite regions within populations. Thus, by determining the length of a set of microsatellites we can identify individuals with a high level of confidence.
We started on this task for my samples last quarter. I am using a set of nine microsatellites that were identified as occurring in Salish Sea harbor seals by Andrew Rothstein during his thesis work. To determine the length of the microsatellites in each sample, each microsatellite must be amplified and documented separately. With approximately 250 samples, this adds up to a lot of PCR! This is made more intimidating by the fact that PCR is inherently finicky. Thus, it is not surprising that there have been some difficulties. See below for one of the first attempts.
PCR #1 - The well at the bottom has the ladder in it. As you
can see, that was all that showed up… Photo by Madelyn
Voelker.
However, we persevered, got a new polymerase, and are beginning to get some success!
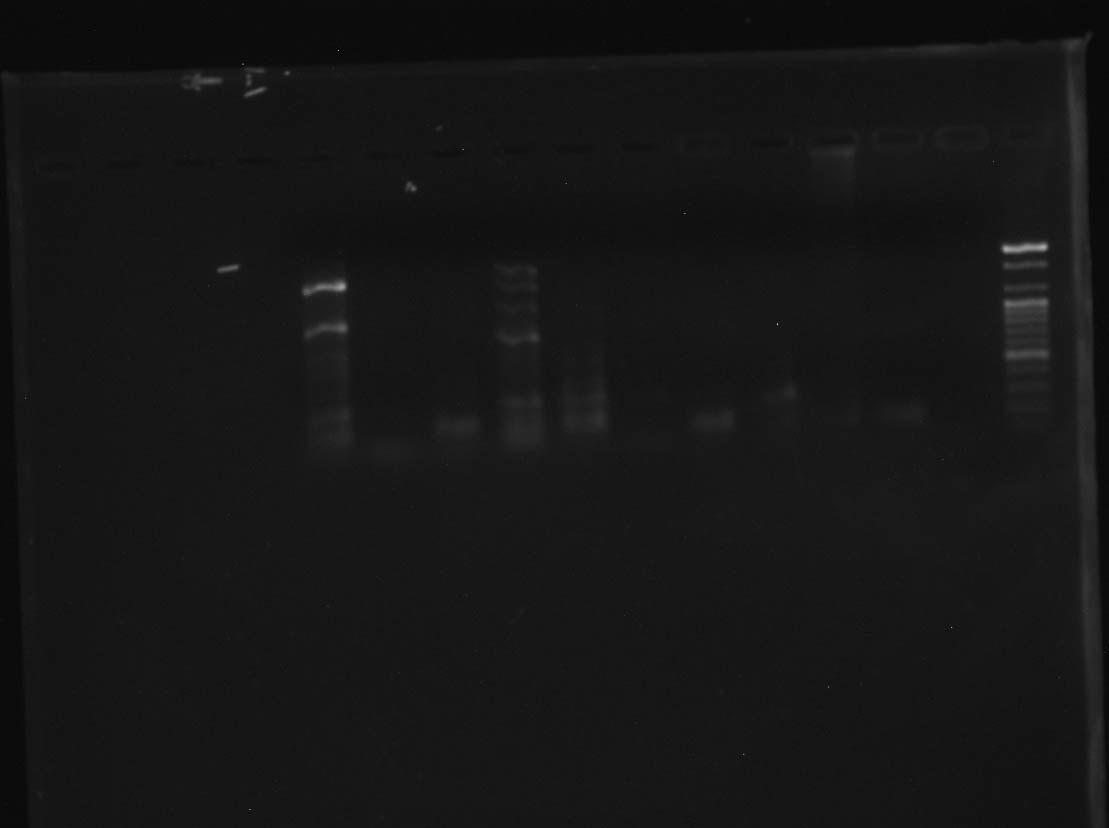
CR #4 - Right to left: Ladder, negative control, positive
control #1, positive control #2, and 9 samples! Photo by Madelyn
Voelker.
As you can see, the majority of samples in this run amplified successfully. Unfortunately, there are multiple bans of DNA showing up in some samples. This is most likely due to non-specific binding of primers. One potential fix for this is to dilute the DNA template before PCR. As a first step in this process, I ran all my samples through the nanodrop, which measures the DNA concentration in the DNA template. My next step will be to use those measurements to calculate how much each sample should be diluted. Then it will be back to PCR!
Marine mammals of the Gulf of Mexico
Rachel Wachtendonk, undergraduate student
1April 2017
Over spring break I went to Cancun, Mexico, on the Yucatan Peninsula. I was able to dive a few times, which allowed me to see some of the amazing sea life; on one dive I saw three sea turtles! This experience made wonder about what marine mammals live in the Gulf of Mexico. I did some research and found that the Gulf of California is home to 30 different cetaceans including killer whales (Orcinus orca), spinner dolphins (Stenella longirostris), Cuvier’s beaked whale (Ziphius cavirostris), and bottlenose dolphins (Tursiops truncatus).



Images: Texas Liberal / Pacific Whale / Whalopedia / National
Geographic
One species from Sirenia is also found in the area, the West Indian manatee (Trichechus manatus).

Image: National Geographic
What I found most interesting is that there are no pinnipeds in this region. There used to be one species of seal, the Caribbean monk seal (Monachus tropicalis). It was determined by NOAA in 2008, after a 5-year review, that the monk seal was extinct. The last sighting of the monk seal was nearly 60 years ago in 1952. These seals became extinct due to being hunted to the point of a population crash. This makes the Caribbean monk seal the first seal species to go extinct due to human influence. There are two more species of monk seal left, the Hawaiian and the Mediterranean monk seals, both are also endangered and at risk of extinction.

Image: The Monachus Guardian
Scientific conference attendance
Raven Benko, undergraduate student
1 April 2017
Part of becoming a scientist involves getting familiar with attending scientific conferences and presenting research. Typically, undergraduates present research in the form of a scientific poster. A scientific poster is much different from what many people may think when they hear “poster presentation”. I discovered this when my roommate entered my room, knowing I was working on a poster for my attendance at the 2017 ALSO Aquatic Sciences Meeting, and was surprised that he didn’t see glitter glue and magic markers. Scientific posters are a professional representation of the research project and include an introduction, methods, results, and conclusion section similar to that of a scientific paper (although extremely abbreviated). I have attended several conferences including the 2016 Salish Sea Ecosystems Conference and the 2016 Larval Fish Conference, but the ASLO Aquatic Sciences meeting was the first conference in which I presented my own research. Below is my completed poster for the conference I attended early March on research from my internship at the J. J. Howard Marine Science Laboratories in Sandy Hook, NJ.
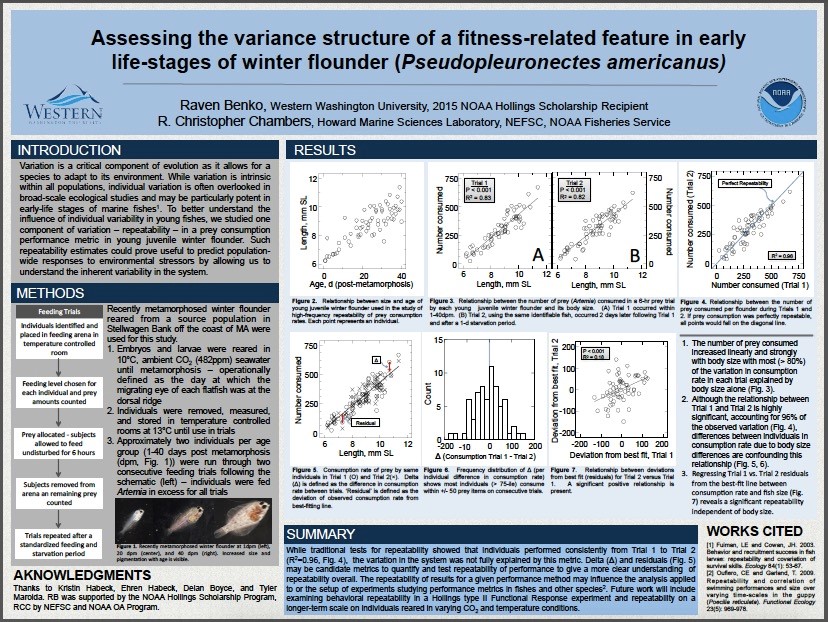
Attending a scientific conference and presenting research can be a rather large and daunting task. Presenters are required to submit an abstract for review and register to attend the meeting either presenting in a poster or oral format. Often times, this is an extremely expensive endeavor. Registration fees, depending on the conference, are generally around $250-$350 for students, not including lodging, flights, or food. The total cost for my attendance at the ASLO meeting was around $3,500 when all travel expenses were included. This conference was, however, located in Honolulu, HI and lasted 5 days therefore it is probably at the upper end of the price point for attending a conference. Many scientists fund these endeavors through their grants or other funding sources and I was fortunate enough to have my attendance at the ASLO meeting supported by the NOAA Hollings Scholarship Program and the WWU Biology Department. I ended up paying very little out of pocket for the amazing opportunity!
The last few weeks I have been preparing to register and submit an abstract to the 2017 Northwest Student Chapter of the Society for Marine Mammalogy conference in Vancouver, B.C. presenting on the work I have done with the WWU Marine Mammal Ecology Lab (in poster format). I am extremely excited by this opportunity as I will be able to learn about current research in marine mammalogy as well as meet students who are researching in the same field. Gaining perspective from Graduate and Ph.D. students as an undergraduate is unbelievably helpful when it comes to preparing for graduate school or entering the workforce. Below is the abstract I submitted for my attendance at the conference:
-
Long-term monitoring reveals evidence of disturbance
threshold at a harbor seal haul-out site in Bellingham,
WA
Interactions between humans and harbor seals (Phoca vitulina) have been steadily increasing, with shoreline construction activities contributing to long-term disturbance of coastal haul-out locations. Responses of marine mammals to construction activities have been well documented, often resulting in temporary or permanent habitat displacement, aversion behavior, stress inducement, and other behavioral changes. Habituation to human disturbance is common in harbor seals - a species that spends large proportions of time hauled-out near the shore. We studied a harbor seal haul-out site in Bellingham Bay located adjacent to a shoreline development project. The area surrounding the haul-out site has experienced increased construction activities including dredging, shoreline armoring, building removal, repaving, and drainage replacement since 2011. Construction is scheduled increase in intensity until project completion around 2025. Long-term monitoring of the haul-out site began in 2007, allowing for comparisons of seal abundances before and after construction. Seal abundances remained constant or slightly increased throughout large-scale construction activities from 2007 until September 2015 when their haul-out structure was removed. Since that removal, seal numbers have substantially decreased – particularly during the pupping season – despite the presence of nearby haul-out replacement structures. Average seal abundance during peak months before 2015 was 32.9±9.08 (n=38 counts) and after 2015 was 5.60±5.49 (n=15 counts). Seals showed evidence of habituation to construction disturbance up to a threshold point and may have tolerated increased construction if the haul-out structures had remained. Our results suggest that when managing shoreline environments, maintenance of historical haul-out structures may reduce the displacement of harbor seals during construction.
I also have one more scientific meeting on the horizon: The 2017 Joint Meeting of Icthyologists and Herpetologists in Austin, TX in July. At this meeting I will be giving my first oral presentation as well as entering in a student researcher competition in which I will be required to write a formal manuscript of my research. The research I will present was in collaboration with Dr. Chris Chambers at the J. J. Howard Marine Science Laboratory (during my internship) studying maternal, paternal, and environmental effects on key hatch characteristics of Atlantic silverside. This will also be an amazing experience to meet lead researchers in the field of fisheries ecology – a subject I may be interested in pursuing for my graduate research. Below is the abstract we submitted for conference attendance:
-
Sources of Variation in Early Life Traits of Atlantic
Silverside, Menidia menidia
Identifying the sources of phenotypic trait variation is critical to determining the potential for adaptive responses to environmental change. We used a quantitative genetics approach to partition the observed variation in early life history (ELH) traits of Atlantic Silverside, Menidia menidia, a forage fish common in inshore waters of the eastern USA. Adults in ripe condition were collected in Sandy Hook Bay, New Jersey and stripped spawned in the laboratory. A paternal half-sibship, hierarchical mating design was implemented and repeated several times during the study. Our largest mating design (D1) used 10 males, each mated to two females with three replicate sets of full-sib offspring. Multiple ELH traits were scored, including egg size; size, age, and survival rate at hatching; and size of larvae at 15 days post-hatch. A smaller design, D2, mated each of 2 males to 2 females (three replicates) and assessed the effects of parentage and two thermal embryo habitats (16 and 22 °C). Design D1 had sufficient power to decompose the variance in each ELS trait into fractions due to mother, father, and genetic variance components, including trait heritability, h2. We found significant differences among females in sizes of eggs (R2=0.59) and larvae at hatching (R2=0.52), and that female average egg size was predictive of average size of larvae at hatching (R2=0.59). The magnitude of the maternal effects lessened with larval age, trait h2 varied between size and age traits, and the multiple environment (D2) design suggested the possibility of genotype x environment interactions.
The work is hard but extremely rewarding and I am excited for the new opportunities ahead!